Import Data¶
The Import Data page is where new samples can be uploaded onto the Genialis Expressions. In order to get started, have your raw data files (typically in .fastq or .fq format), preferably compressed (zip, gz, etc.) on hand, or your BaseSpace login credentials to ensure a smooth and easy upload of your samples to Genialis Expressions.
The Import Data page consists of two cards:
Import Files
Collection Details
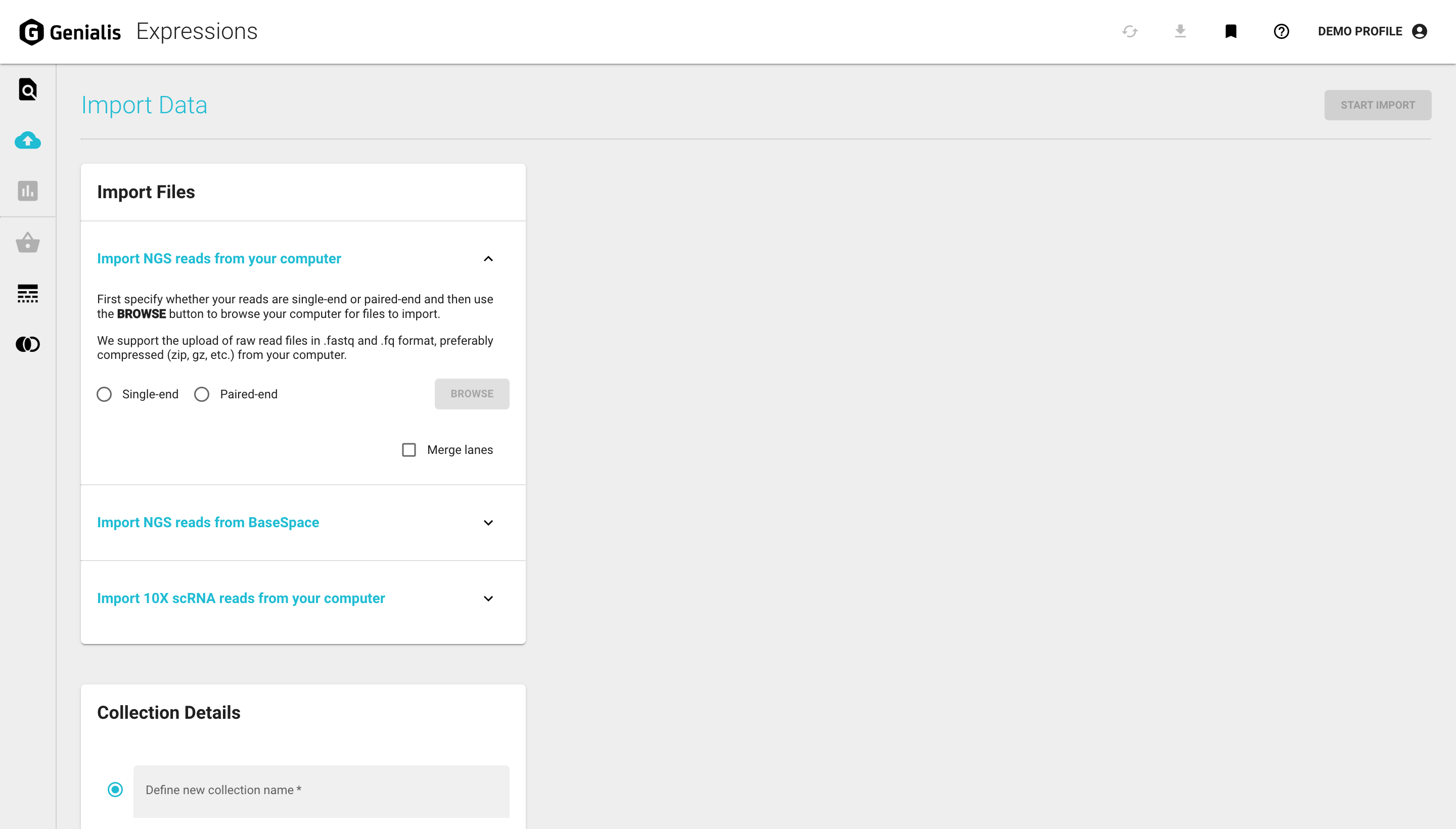
You can import your single-end or paired-end raw data files from your local drive, your BaseSpace account, or import 10X scRNA reads from your local drive.This process is described in detail below.
All your new samples are automatically put into a collection for easier data maintenance. A collection is simply an arbitrary group of samples that belong together. In the Collection Details card you will need to provide a meaningful name for your new collection. You can also assign the newly imported samples to an existing collection. You can read more about data organization on Genialis Expressions in the Search for Data article.
Import NGS reads from your computer¶
If you are importing data from your computer, navigate to the ‘Import NGS reads from your computer’ section of the Import Files card.
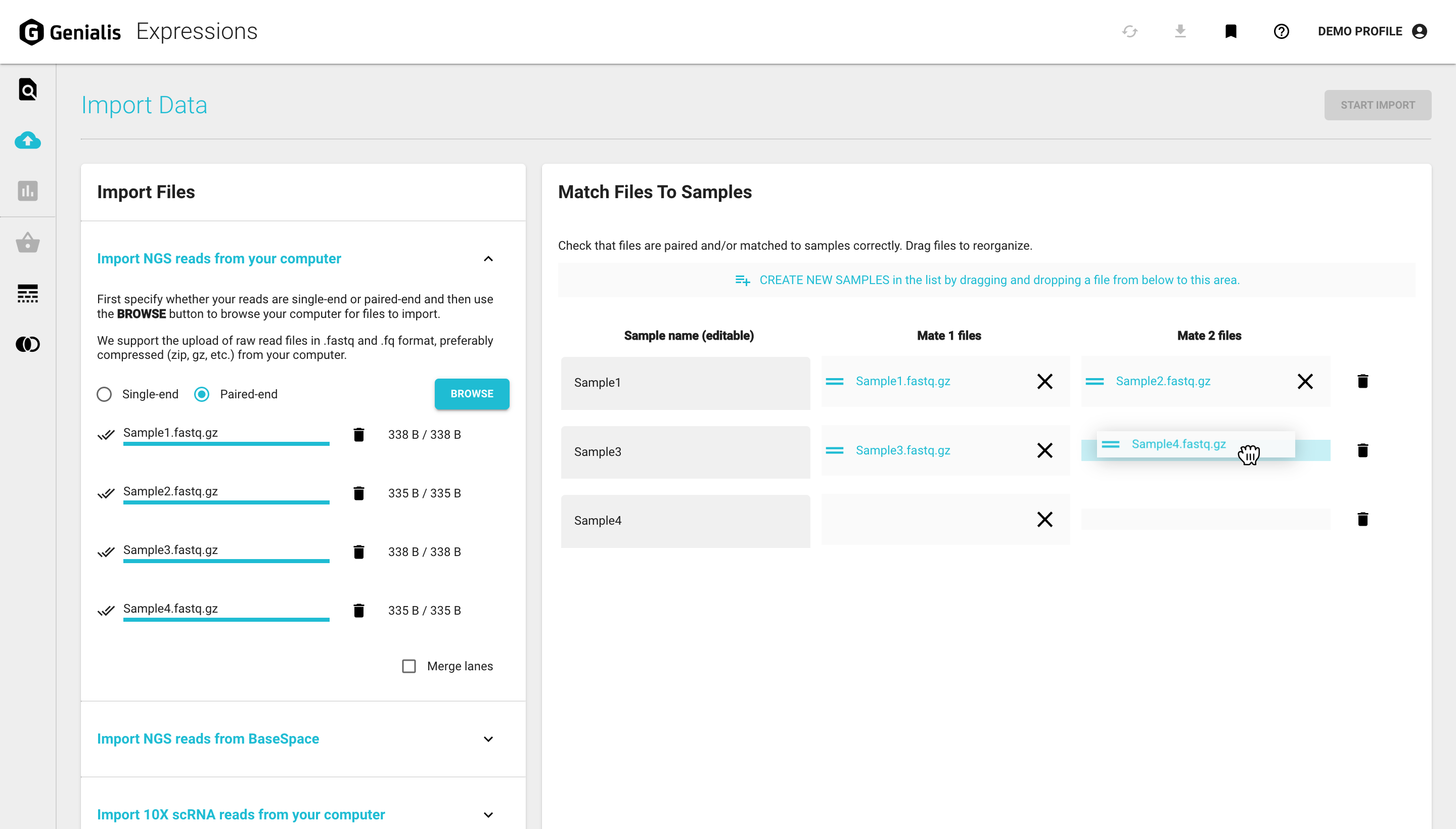
Begin by selecting whether your sample is single-end or paired-end. Click on ‘BROWSE’, and a computer system window will appear where you can locate your files on your computer.
Once you select the files to import, click ‘OPEN’ and you will see them appear in the Import Files card (where you can monitor the file upload progress).
The files will also show up in a table in a new card called Match Files to Samples on the right side of the page. This is where you should check that your files are paired and/or matched to the correct samples. If they are not, drag and drop the files to reorganize or create a new sample by dragging and dropping an imported file to the ‘ADD SAMPLE TO THE LIST’ sign above the table.
If your samples are multi-lane, you can also upload them using our import page. Just select the ‘Merge lanes’ option and be sure to double check that your multi-lane samples are arranged correctly before you import them.
Once your files are set up, this is the perfect opportunity to change your samples names to be more descriptive (e.g. WT-DrugTreated-Rep1). You can do this by updating the name within the ‘Sample name (editable)’ column. This is the only time you’ll need to operate with files, your future work should mostly concern samples and collections. Click here to read about these concepts.
Once completed, click ‘START IMPORT’ to start the import process. After the import is finished, you will be redirected to the Collection Details page, where you can manage the collection to which the new samples now belong and run further bioinformatic tools and pipelines.
Import NGS reads from BaseSpace¶
If you are importing data from your BaseSpace account, be sure you have your Illumina BaseSpace credentials. Now navigate to the ‘Import NGS reads From BaseSpace’ section of the Import Files card.
After clicking on ‘SIGN IN TO BASESPACE’ and logging in to your BaseSpace account, you will receive a prompt indicating that Genialis Expressions is requesting permission to access your BaseSpace account. You’ll only be asked for permission the first time you log in to your BaseSpace account from Genialis Expressions. After granting this permission, you will be redirected back to the Import Files card. You will now be able to browse and select the samples you wish to import. After selecting your samples, click ‘IMPORT SAMPLES’.
Once completed, click ‘START IMPORT’ to start the import process. You will now be redirected to the Collection Details page, where you can manage the collection to which the new samples now belong or run bioinformatic tools and pipelines.
Import data from other sources¶
We can also help import your data from other sources including importing directly from BaseSpace, using ReSDK (an open-source, Python-based application programming interface developed by Genialis), file transfers via FTP, importing data from the Gene Expression Omnibus (GEO) or the Sequence Read Archive (SRA) database. Please contact us at support@genialis.com to arrange this service.