Genialis Supermodel Platform¶
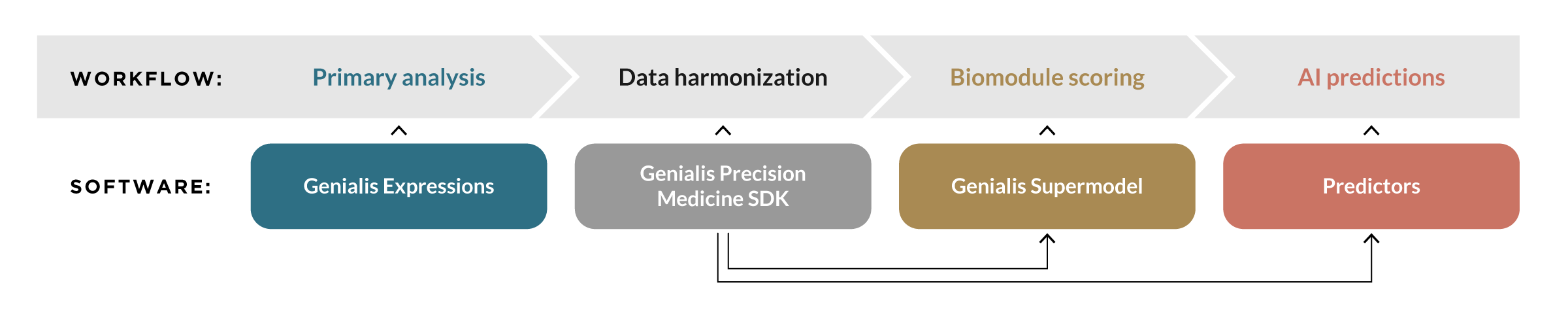
Genialis Supermodel creates therapeutic intelligence by interpreting oncology biospecimens with biomodule scores. The workflow has four main phases:
Primary analysis analyzes raw RNA sequencing data and produces gene expression profiles
Data harmonization includes data quality assurance, data pre-processing, and batch effect detection and mitigation
Biomodule scores are computed with the Genialis Supermodel; biomodules are algorithmic abstractions of biological pathways, gene networks, or specific biological states
AI predictions are made by applying existing predictors (AI models) to the biomodule scores.
The Genialis Supermodel Platform includes four key software components that support this workflow:
Genialis Expressions is a robust and scalable cloud software that captures analysis metadata and handles primary processing of sequencing data via a suite of validated pipelines
Genialis Precision Medicine SDK prepares data for machine learning using data normalization, a system of preprocessors, and a framework for batch effect detection and removal
Genialis Supermodel transforms harmonized sequencing data into a low-dimensional biological space using biomodules
Predictors are AI models developed to predict response, prognosis, or other clinically relevant insights; these are trained by the Supermodel licensees or contracted and licensed directly from Genialis
Each of the software components comes with an API allowing deployment of all or a selection of these components within existing software architectures.
Below, the components are introduced with some more detail. The complete documentation is accessible from the Table of Contents.
1. Genialis Expressions: Scalable, Secure, and Reproducible Bioinformatics¶
Genialis Expressions provides a robust pipeline infrastructure built on Genialis’ open source Resolwe dataflow engine and deployed on a scalable, cost-effective, and secure microservices architecture. It contains validated bioinformatic pipelines for diverse data types, including RNA-Seq, DNA-Seq (WES/WGS), ChIP-seq, ATAC-seq, and WGBS. The system complies with strict regulatory and cybersecurity standards (HIPAA, GDPR, OWASP, CIS, NIST).
RNA-Seq workflows support both traditional and pseudo-alignment quantification (STAR, Salmon), while proprietary custom workflows enable variant detection from RNA-derived data. All pipelines are production-hardened and modular, ensuring consistent annotation and QC across metadata dimensions.
Genialis Expressions offers both programmatic (Python-based ReSDK) and graphical access via a browser-based user interface, enabling data scientists and translational scientists to work efficiently with stable, consistently processed and annotated datasets.
2. Genialis Precision Medicine SDK¶
Genialis Precision Medicine SDK (GPM) is a production-ready Python SDK for preparing sequencing data for machine learning (ML) applications and for training new predictors or components of the Supermodel.
Data harmonization begins with data ingestion and quality control. Next, systematic noise in data (distribution drift, batch effect) can be detected and mitigated. Finally, Genialis’ patent-pending (US20240233874A1) preprocessing system transforms raw gene expression data into harmonized, machine learning-ready input features. One of the core components of this framework is rnanorm, a scikit-learn–compatible Python package that supports both within-sample and between-sample normalization strategies. It corrects for factors like library size and gene length while ensuring comparability across samples. Application-specific preprocessors are available to transform input data for individual predictors. This proprietary system of preprocessors is extensible, featuring a growing collection of tissue- and model-specific preprocessors with built-in functionality for batch effect detection and correction.
GPM also implements strong ML best practices for high-dimensional, low-sample-size datasets (HDLSS), common in precision medicine applications. GPM can be used to train new biomodules to be included in the Supermodel, or new predictors to be used at various stages of drug development.
3. Genialis Supermodel: interpretable encoding of biology through biomodules¶
The core component of the Genialis Supermodel is a growing library of pretrained biomodules. Biomodules are algorithmic abstractions of biological phenomena that transform high-dimensional transcriptomic data into interpretable, biologically grounded features. Each biomodule functions as an axis in a multidimensional “biological space,” enabling dimensionality reduction and improved generalizability. Each biomodule produces a numerical score that reflects activity in biological pathways, gene networks, or specific biological states. Genialis Supermodel can be provided by Genialis as a service, or deployed on an external infrastructure and run directly by licensees.
Biomodules are developed using reusable ML pipelines that standardize their definition and construction. A defined maturity process governs the progression of biomodules from early discovery to deployment.
4. Predictors: AI Models for Clinical and Translational Insight¶
Genialis predictors are modular, AI-driven models engineered to predict biological traits (e.g., MSI status), clinical outcomes (e.g., survival), or therapeutic responses (e.g., to KRAS inhibitors). These predictors are developed using standardized pipelines and FDA-guided Good Machine Learning Practices that emphasize reproducibility, interpretability, and regulatory readiness. Each model includes versioned components and is validated for performance, rigor, and traceability. Predictors can be deployed in multiple formats, including Docker containers, APIs, command-line tools, or Python packages, and support batch processing from standard data inputs such as CSV and FASTQ files.
See this krasID White Paper for an example of a predictor. Genialis can be contracted to develop custom predictors. Alternatively, users of the Genialis software can leverage the GPM code library, which provides adapted machine learning methods and best practices tailored for small datasets commonly encountered in precision medicine.